In Vitro Drug Sensitivity of Ureaplasma Species and Mycoplasma Hominis and the Mechanism of Resistance to Quinolones: A Single-center Retrospective Study
-
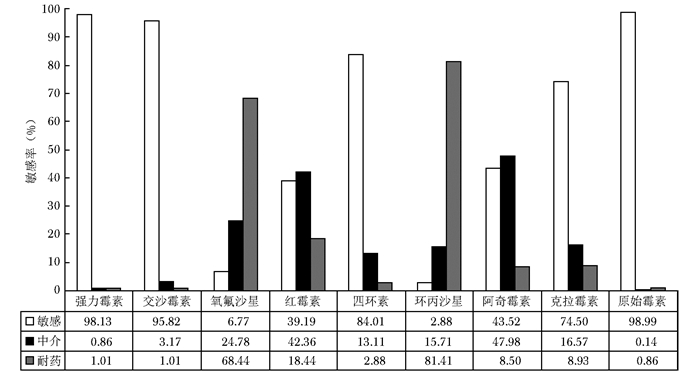
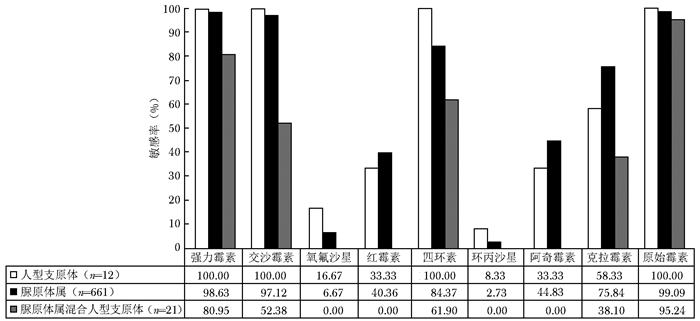
摘要:目的 分析脲原体属和人型支原体体外药物敏感性特点, 并分析其对喹诺酮类药物耐药的相关机制。方法 回顾性总结北京协和医院2012年9月至2017年4月通过体外培养方法检测出的脲原体属和人型支原体标本, 结合患者临床资料和菌种鉴定结果分析其体外药物敏感性特点。针对喹诺酮类药物耐药特点, 采用普通PCR法扩增目的基因并测序, 将测序结果翻译成蛋白质序列, 与NCBI数据库中参考序列进行比对, 检测DNA促旋酶(GyrA/GyrB)和拓扑异构酶Ⅳ(ParC/ParE)序列的突变情况。结果 脲原体属混合人型支原体的体外敏感性总体低于脲原体属或人型支原体单独体外敏感性; 除大环内酯类外, 脲原体属对喹诺酮类、四环素类、交沙霉素、原始霉素、强力霉素的敏感性普遍低于人型支原体, 但差异无统计学意义(P均>0.05)。女性患者体内分离得到的脲原体属对阿奇霉素、红霉素、克拉霉素、氧氟沙星的体外敏感性较男性低(P均<0.05)。微小脲原体对大部分抗菌药物的敏感性高于解脲脲原体, 尤以对四环素的敏感率差异最大(98.48%比72.73%, 差值25.8%, P<0.05)。在GyrA、GyrB、ParC和ParE的序列比对中, 共发现21个突变位点, 其中ParC S83L突变占96.22%(51/53), 为最主要突变位点; 其余为ParC A136T突变、ParE R448K突变、GyrA L176F和ParC S83L联合突变等以及6个目前尚未报道过的突变位点, 即ParC的L540F、R718W、Q767E、S789N、M828I和I831T突变。结论 脲原体属和人型支原体的体外药物敏感性与感染种属有关, 脲原体属的体外敏感性还与菌种、患者性别有关。对脲原体属喹诺酮类耐药的机制研究表明, 单独ParC S83L突变是脲原体属对喹诺酮类耐药的主要机制, 而新发现6个突变位点可能的作用机制仍有待深入研究。Abstract:Objective The aim of this study was to summarize and analyze the in vitro antimicrobial susceptibility of Ureaplasma species and Mycoplasma hominis and the mechanisms responsible for resistance to quinolones.Methods The clinical data of Ureaplasma species and Mycoplasma hominis detected by in vitro culture from September 2012 to April 2017 in Peking Union Medical College Hospital were retrospectively collected and analyzed; their characteristics of antimicrobial susceptibility were also analyzed combined with the information about the patients and species identification. According to the drug-resistance characteristics of quinolone, the target genes were amplified and sequenced by PCR, and the sequencing results were translated into protein sequences. The mutations in DNA gyrase(GyrA/GyrB) and topoisomerase Ⅳ (ParC/ParE) that were related to quinolones resistance were detected by comparing sequences in the NCBI database.Results In vitro sensitivity to antibiotics of Ureaplasma species mixed with Mycoplasma hominis was significantly lower than that of Ureaplasma species or Mycoplasma hominis alone. Compared to Mycoplasma hominis, Ureaplasma species was less susceptible to quinolones, tetracycline, josamycin, and primycin, except for macrolides, but the difference was not statistically significant (all P > 0.05). In addition, the susceptibility of Ureaplasma species to azithromycin, erythromycin, clarithromycin, and ofloxacin in female patients was lower than that in male patients (all P < 0.05). Ureaplasma parvum were more susceptible than Ureaplasma urealyticum to most antibiotics, especially tetracycline (98.48% vs. 72.73%; 25.75% discrepancy, P < 0.05). Moreover, twenty-one mutations from sequences of GyrA, GyrB, ParC, and ParE were determined. The mutation in ParC with S83L substitution was the most frequent, 96.22% (51/53); mutations of A136T substitution in ParC, R448K substitution in ParE, and L176F in GyrA combined with S83L in ParC were also detected. This study also found six novel mutations that have not been reported:L540F, R718W, Q767E, S789N, M828I, and I831T amino acid substitutions in ParC protein.Conclusions The in vitro antimicrobial susceptibility of Ureaplasma species or Mycoplasma hominis is associated with genus, and the in vitro sensitivity of Ureaplasma species to antibiotics is correlated with species and genders of patients. Sole S83L substitution in ParC might be the major mechanism of resistance to quinolones of Ureaplasma species, while the possible function of the six novel mutations remains further studies.
-
中枢神经系统感染性疾病是临床重要的急危重症,在世界范围内具有较高的发病率和病死率[1]。早期识别致病病原体并给予针对性治疗,是改善患者预后的关键[2]。尽管如此,采用传统的病原学检测技术进行中枢神经系统感染性疾病诊断,仍存在超过半数的感染无法明确病原体[3]。宏基因组高通量测序(metage-nomic next-generation sequencing,mNGS) 技术可非靶向检测临床标本中存在的细菌、真菌、病毒和寄生虫等病原体核酸,具有广谱和灵敏度高的特点,其临床应用越来越广泛。目前,国内外相关研究也肯定了mNGS技术在中枢神经系统感染性疾病中的诊断价值[2, 4-7]。
2017年,中国医学科学院北京协和医院检验科团队开始探索建立mNGS检测技术流程,并于2022年正式投入临床应用。不同于第三方实验室,该检测平台的报告解读基于技术解读和临床解读两个层面,由从事生物信息学、临床感染、临床微生物学等专业人员合作完成。本研究基于该检测平台,对临床诊断为中枢神经系统感染性疾病患者的脑脊液mNGS检测结果进行回顾性分析,评价其在中枢神经系统感染性疾病诊断中的价值。
1. 资料与方法
1.1 临床资料
本研究为回顾性诊断性试验,研究对象为2022年4—12月于中国医学科学院北京协和医院就诊的患者。纳入标准:(1)患者最终临床诊断为中枢神经系统感染性疾病,参考《北京脑炎协作组脑炎诊断标准》[8];(2)行脑脊液mNGS检测。排除标准:脑脊液细胞学证实为中枢神经系统恶性肿瘤或已确诊为自身免疫性脑炎的患者。
本研究已通过中国医学科学院北京协和医院伦理审查委员会审批(审批号:I-23ZM0049),并豁免患者知情同意。
1.2 研究方法
登录医院信息系统,查阅并收集平台投入临床使用以来患者脑脊液mNGS检测报告结果,同时收集患者的相关临床信息、脑脊液常规、生化及病原学检查等资料。
脑脊液mNGS检测由中国医学科学院北京协和医院感染性疾病宏基因组检测平台开展。具体方法:采集患者1~2 mL脑脊液标本送检,同步进行DNA和RNA核酸提取,随后进行文库构建,并在Illumina平台进行测序,通过数据分析(广州微远基因生物公司提供平台及协和自建分析流程),获得微生物检出及比对结果。
mNGS的结果解读流程分为技术解读和临床解读两个层面[9],解读人员由多名具有丰富mNGS报告解读经验的人员构成,包括1名具有生物信息学教育背景的人员、1名具有临床检验诊断学及生物信息学专业知识的人员、1名具有微生物学及感染病学专业知识的人员。
以患者的临床诊断作为金标准,相关定义如下:(1)真阳性:mNGS检出病原体,且与患者的临床诊断符合;(2)假阴性:mNGS未检出病原体,但患者的临床诊断为中枢神经系统感染。
1.3 样本量估算
根据既往研究数据[2-3],采用样本量计算公式:$ n=p(1-p)\left(\frac{z_{1-\alpha / 2}+z_{1-\beta}}{p-p_0}\right)^2$,取双侧检验,在α=0.05,β=0.1的条件下,计算本研究所需最小样本量为19例。
1.4 统计学处理
采用SPSS 26.0统计软件对数据进行分析,符合正态分布的计量资料以均数±标准差表示,偏态分布的计量资料以中位数(四分位数)表示,计数资料采用频数(百分数)表示。
2. 结果
2.1 患者临床资料
共入选39例符合纳入和排除标准的中枢神经系统感染性疾病患者,其中男性22例(56.4%),女性17例(43.6%),平均年龄(50.5±15.9)岁。主要临床表现为发热、头痛、意识障碍、癫痫发作、肢体麻木/无力、大小便障碍等(表 1)。急性病程22例(56.4%),亚急性/慢性病程17例(43.6%)。患者入院后均完善腰椎穿刺检查,并送检脑脊液常规和生化检查,脑脊液白细胞计数为148(62,221)×106 /L,蛋白为1.46(0.89,2.45)g/L,葡萄糖为3.2(2.0,4.6)mmol/L,氯化物为(117.9±9.1)mmol/L。
表 1 39例中枢神经系统感染者的临床表现[n(%)]临床表现 数值 发热 27(69.2) 头痛 19(48.7) 呕吐 4(10.3) 意识障碍 17(43.6) 癫痫发作 5(12.8) 肢体麻木/无力 4(10.3) 精神行为异常 2(5.1) 认知功能下降 2(5.1) 大小便障碍 2(5.1) 视物模糊 1(2.6) 听力下降 1(2.6) 语言障碍 1(2.6) 吞咽障碍 1(2.6) 行走不稳 1(2.6) 颅内占位 2(5.1) 皮疹 5(12.8) 2.2 脑脊液mNGS检测结果与临床验证
在39例患者中,10例(25.6%)脑脊液mNGS报告结果为阴性;29例(74.4%)检出疑似致病病原体特异性序列,其中11例(37.9%)检出细菌,13例(44.8%)检出病毒,3例(10.3%)检出真菌,1例(3.5%)检出细菌及病毒,1例(3.5%)检出真菌及病毒。检出细菌以结核分枝杆菌最为常见(6例),其次为金黄色葡萄球菌(2例)、肺炎克雷伯菌(1例)、屎肠球菌及肺炎克雷伯菌混合感染(1例)、链球菌属及表皮葡萄球菌(各1例)。检出病毒以水痘-带状疱疹病毒最为常见(9例),其次为EB病毒(3例,其中1例同时检出新型隐球菌及EB病毒)、巨细胞病毒(2例,其中1例同时检出链球菌属及巨细胞病毒)及肠道病毒71型(1例)。检出真菌以新型隐球菌最为常见(2例),其次为白色念珠菌(1例)及烟曲霉(1例)。
29例阳性病例脑脊液mNGS检测结果均与临床诊断相符,真阳性率为74.4%。阳性病例脑脊液mNGS病原体检测结果与临床验证情况详见表 2。在检出结核分枝杆菌的6例患者中,除1例患者于送检当日死亡外,其余5例患者临床诊断均为结核性脑膜炎/脑膜脑炎,并接受抗结核治疗。在此6例患者中,4例送检脑脊液X-pert® MTB/XDR检测,仅1例结果为阳性;2例送检脑脊液结核分枝杆菌核酸聚合酶链式反应(polymerase chain reaction,PCR)检测,结果均为阴性。在mNGS检出细菌阳性(含细菌及病毒混合检出1例)的其他患者中,除1例患者(检出金黄色葡萄球菌阳性)入院后死亡未留取病原学标本外,其余5例检测结果均与血培养/脑脊液/硬膜外脓肿脓液培养结果一致。在检出病毒的患者中,除2例细菌及真菌混合感染外,其余13例临床诊断均为病毒性中枢神经系统感染,并接受抗病毒治疗,2例脑脊液病毒核酸PCR检测阳性。在mNGS检出真菌(含真菌及病毒混合检出1例)的患者中,仅1例脑脊液培养为阳性,2例检出新型隐球菌的病例其脑脊液隐球菌抗原均为阳性,1例检出烟曲霉的病例其血曲霉半乳甘露聚糖(galactomannan,GM)试验为阳性。
表 2 29例阳性病例脑脊液mNGS病原体检测结果与临床验证情况mNGS检测结果 临床验证方法 细菌感染(12例) 结核分枝杆菌(6例) 临床诊断结核性脑膜炎/脑膜脑炎并接受抗结核治疗(5例) 涂片阳性肺结核(1例) 脑脊液X-pert® MTB/XDR检测阳性(1例) 金黄色葡萄球菌(2例) 血培养阳性(1例) 脑脊液白细胞明显升高且以多个核细胞为主(2例) 肺炎克雷伯菌(1例) 血培养阳性(1例) 屎肠球菌及肺炎克雷伯菌混合感染(1例) 硬膜外脓肿脓液培养阳性(1例) 链球菌属(1例) 脑脊液培养阳性(1例) 表皮葡萄球菌(1例) 脑脊液培养阳性(1例) 病毒感染(13例,除外2例混合感染) 水痘‐带状疱疹病毒(9例) 临床诊断病毒性脑膜炎/脑膜脑炎/脑脊髓炎并接受抗病毒治疗(9例) 疱疹(5例) EB病毒(2例) 脑脊液EB病毒核酸PCR检测阳性(1例) 临床诊断病毒性脑膜炎/脑炎并接受抗病毒治疗(2例) 巨细胞病毒(1例) 脑脊液巨细胞病毒核酸PCR检测阳性(1例) 临床诊断病毒性脑膜炎/脑炎并接受抗病毒治疗(1例) 肠道病毒71型(1例) 临床诊断病毒性脑膜炎并接受抗病毒治疗(1例) 真菌感染(4例) 新型隐球菌(2例) 脑脊液隐球菌抗原阳性(2例) 脑脊液培养阳性(1例) 烟曲霉(1例) 血曲霉半乳甘露聚糖试验阳性(1例) 白色念珠菌(1例) 临床诊断念珠菌性中枢神经系统感染(1例) mNGS:宏基因组高通量测序;PCR:聚合酶链式反应 10例(25.6%)患者的mNGS检测结果为假阴性,根据其临床表现、脑脊液常规及生化结果等判断,临床诊断为中枢神经系统病毒感染4例,考虑细菌感染4例,无明确病原学指向2例。此10例患者的脑脊液临床常规病原学检查无阳性结果回报。
38例患者进行了脑脊液临床常规病原学检查(包括脑脊液涂片、培养、病原体抗原及PCR核酸检测)。5例(13.2%)脑脊液/硬膜外脓肿脓液培养阳性,其中细菌4例,真菌1例。1例脑脊液X-pert® MTB/XDR检测阳性,2例脑脊液病毒核酸PCR检测阳性,2例脑脊液隐球菌抗原检测阳性。整体脑脊液临床常规病原学检查的阳性率为23.7%(9/38)。
39例中枢神经系统感染患者中,16例临床诊断为细菌感染(包括7例结核性脑膜炎/脑膜脑炎、5例化脓性脑膜炎、3例脑脓肿及1例硬膜外脓肿),17例临床诊断为病毒感染(包括8例脑膜炎、4例脑炎、3例脑膜脑炎、1例脑脊髓炎及1例脑脊髓膜炎),4例临床诊断为真菌感染(包括2例隐球菌性脑膜炎、1例烟曲霉及1例念珠菌性中枢神经系统感染),2例临床无明确病原学指向。mNGS病原体检测结果与临床诊断比较详见表 3。对于结核性脑膜炎/脑膜脑炎,mNGS诊断的灵敏度、特异度分别为85.7%、100%;对于其他中枢神经系统细菌感染,mNGS诊断的灵敏度、特异度分别为66.7%、100%;对于中枢神经系统病毒感染,mNGS诊断的灵敏度、特异度分别为76.5%、86.4%;对于中枢神经系统真菌感染,mNGS诊断的灵敏度、特异度均为100%。
表 3 mNGS病原体检测结果与临床诊断比较[n(%)]mNGS检测结果 临床诊断 细菌(n=9) 结核(n=7) 病毒(n=17) 真菌(n=4) 检出对应致病病原体(n=29) 6(66.7) 6(85.7) 13(76.5) 4(100) 未检出对应致病病原体(n=10)* 3(33.3) 1(14.3) 4(23.5) 0(0) mNGS: 同表 2;*包含2例临床无明确病原学指向的中枢神经系统感染病例 3. 讨论
本研究通过回顾性分析发现,39例临床诊断为中枢性神经系统感染性疾病患者中,29例通过mNGS检出疑似致病病原体,且均与临床诊断一致,真阳性率为74.4%,显著高于常规病原学检查结果(74.4%比23.7%)。
中枢神经系统感染性疾病作为临床急危重症,对致病病原体的早期识别和治疗是改善患者预后的关键[2]。脑脊液涂片及培养作为传统的病原学检测方法,不仅阳性率低,且培养耗时较长,不利于早期诊断。尽管脑脊液病原学抗原检测及基于PCR的核酸检测在临床逐渐推广,但其联合涂片及培养检测的阳性率未超过50%[2-3, 5]。为提高检测的阳性率,缩短检测时间,有研究引入质谱技术,无需进行培养即可直接检出脑脊液中的病原体[10],但目前该技术在国内外应用的报道较少,且脑脊液中需含有较高的菌量方可检出[11]。mNGS技术相较于传统检测方法,灵敏度更高且耗时更短,有助于临床疑难及急危重感染性疾病病原体的快速识别和诊断。
目前,国内外相关研究对mNGS技术在中枢神经系统感染性疾病诊断中的价值进行了探讨。国外一项纳入213例中枢神经系统感染性疾病患者的多中心、前瞻性研究显示,mNGS技术在中枢神经系统感染性疾病中的病原体阳性检出率为57%,在检测病毒、细菌及结核分枝杆菌感染方面优于传统检测手段[5]。国内研究显示,mNGS技术对中枢神经系统感染性疾病的病原体阳性检出率为41%~47%[7, 12-13]。尽管mNGS技术在结核分枝杆菌的检出方面存在一定局限性(核酸提取难度大,导致检出序列少或出现假阴性)[14],但本研究依然显示出脑脊液mNGS检测在结核性脑膜炎/脑膜脑炎诊断中的优势,在7例临床表现及脑脊液检测诊断为结核性脑膜炎/脑膜脑炎的病例中,6例mNGS检出结核分枝杆菌,灵敏度为85.7%。而在此7例患者中,仅1例(14.3%) 痰抗酸染色为阳性,1例(14.3%)结核分枝杆菌PCR检测为阳性,mNGS检测的阳性率明显高于临床常规检测手段。且脑脊液mNGS检出结核分枝杆菌亦具有极好的特异性,这与国外文献报道一致[5, 15]。
mNGS检出的29例阳性病例均与临床诊断相符,并可通过病原学检查(脑脊液涂片、培养、病原体抗原及PCR检测)验证或接受相应的抗微生物治疗。但在mNGS检出病毒的病例中,2例确诊为中枢神经系统细菌/真菌感染的病例同时检出了超过10条序列的病毒,且均为条件致病病毒(EB病毒及巨细胞病毒各1例),考虑可能为机体的潜伏感染。
本研究中,mNGS技术检出病原体的真阳性率为74.4%,高于国内外文献报道,此结果与本实验室对于报告解读流程的改进有关。mNGS技术的检测流程复杂,检测结果易受多方面因素的影响,实验室及试剂中的背景微生物、腰椎穿刺采样过程中皮肤定植菌的污染、采样及送检过程中环境微生物的污染等均对检测报告的解读存在干扰。目前,国内mNGS检测分为院内检测和第三方实验室检测两种方式。自2017年起,中国医学科学院院北京协和医院检验科团队从临床需求出发,先后建立了mNGS的湿实验流程、生物信息分析流程、背景核酸数据模型,形成了相对规范的mNGS检测流程[16-18]。在临床应用前,本团队结合已发表的专家共识[14, 19],与急诊科、重症医学科、感染内科、神经科等多个科室的医师团队共同撰写报告解读流程文件[9, 20]。在报告解读过程中,通过背景核酸数据模型过滤试剂、采样、环境中的背景微生物,同时结合患者的临床病史、脑脊液常规检测结果、病原学检测结果、影像学检查结果等综合判断检出结果与临床的一致性,最大可能避免假阳性报告的签发。同时,对于临床疑似中枢神经系统感染的患者标本,在首次检测结果未检出明确病原体的情况下,检验医师在查阅临床病例资料后指导调整湿实验流程,如提高破壁强度以提升真菌、结核分枝杆菌的核酸提取效率[21-22],增加去宿主流程以降低人源基因的比例,提升检测的真阳性率[23-24]。对于疑难病例,实验室则通过反复临床沟通、多学科会诊等策略与临床医师合作,尽可能解决临床问题。
本研究局限性:共纳入39例中枢神经系统感染性疾病病例,样本量相对较小,可能影响结果的外推性。mNGS技术作为新兴的检测技术,相较于传统检查手段,目前费用昂贵,同时对检测人员和解读人员的要求也更高,在一定程度上限制了其临床应用。随着技术的不断进步,通过降低成本费用、简化检测流程、缩短周转时间,相信mNGS技术可得到更广泛的应用。
综上,在中枢神经系统感染性疾病病原体检测方面,脑脊液mNGS检测相较于现有临床常规病原学检测手段具有更高的真阳性率,能够较为高效、快速、准确地获得致病病原体的信息,对于中枢神经系统感染性疾病的早期诊断及治疗、改善患者临床预后具有重要价值。
利益冲突 无刘亚丽、张文娟、王洁对本文同等贡献 -
引物名称 引物序列 退火温度(℃) 目的基因长度(bp) gyrA-1 5′-TTGCTGCTTTCGAAAACGG-3′ 50 336 gyrA-2 5′-CTGATGGTAAAACACTTGG-3′ gyrB-1 5′-GAAAATAGACGTGGTCG-3′ 55 1668 gyrB-2 5′-TTCGAATATGACTGCCATC-3′ gyrB-3 5′-CCTGGTAAATTAGCTGACTG-3′ 55 1249 gyrB-4 5′-CTGATGGTAAAACACTTGG-3′ parC-1 5′-ACGCAATGAGTGAATTAGG-3′ 55 309 parC-2 5′-CACTATCATCAAAGTTTGGAC-3′ parC-3 5′-GCTAAAACGCTACAAGAACG-3′ 55(UP)/48(UU) 1318(UP)/1324(UU) parC-4 5′-CAACGTTGGCATAAATTGG-3′ parE-1 5′-ATGGGCGGAAAATTAACGC-3′ 55 313 parE-2 5′-CTTGGATGTGACTACCATCG-3′ UP:微小脲原体;UU:解脲脲原体 表 2 不同性别间脲原体属对9种抗菌药物体外敏感性差异比较
(%, N=661) 抗菌药物 男性(n=71) 女性(n=590) 敏感 中介 耐药 敏感 中介 耐药 强力霉素 98.57 1.43 0.00 98.64 0.68 0.68 交沙霉素 97.14 1.43 1.43 97.11 2.55 0.34 氧氟沙星 12.86 44.29 42.86 5.93 23.05 71.02 红霉素 60.00 30.00 10.00 38.03 44.48 17.49 四环素 90.00 7.14 2.86 83.70 14.09 2.21 环丙沙星 2.86 18.57 78.57 2.71 15.76 81.53 阿奇霉素 62.86 28.57 8.57 42.69 51.02 6.29 克拉霉素 84.29 8.57 7.14 74.83 17.52 7.65 原始霉素 100.00 0.00 0.00 98.98 0.17 0.85 表 3 微小脲原体与解脲脲原体体外药物敏感性比较
(%) 抗菌药物 微小脲原体(n= 66) 解脲脲原体(n=11) 敏感 中介 耐药 敏感 中介 耐药 强力霉素 98.48 1.52 0.00 90.91 0.00 9.09 交沙霉素 98.48 1.52 0.00 100.0 0.00 0.00 氧氟沙星 10.61 33.33 56.06 27.27 9.09 63.64 红霉素 65.15 30.30 4.55 72.73 18.18 9.09 四环素 98.48 1.52 0.00 72.73 18.18 9.09 环丙沙星 3.03 16.67 80.30 0.00 9.09 90.91 阿奇霉素 69.70 25.76 4.55 63.64 27.27 9.09 克拉霉素 95.45 0.00 4.55 81.82 9.09 9.09 原始霉素 98.48 1.52 0.00 100.00 0.00 0.00 表 4 喹诺酮类药物耐药相关突变筛查结果
标本编号 氧氟沙星
(μg/ml)环丙沙星
(μg/ml)氨基酸序列改变 GyrA ParC ParE 微小脲原体 支3/4/9/31/35/38/43/44/45/46/47/53/54/55/57/59/60/62/66/67/80/87/90/92/98/101;尿20/55/64/96(n=30) R(≥4) R(≥2) S83L 支28/33/41/49/63/95/100;尿10/38
(n=9)I(>1且<4) R(≥2) S83L 支34(n=1) R(≥4) R(≥2) S83L, M105I 支56;尿56(n=2) S(≤1) R(≥2) S83L 支71;尿28(n=2) R(≥4) R(≥2) S83L, R718W 支81(n=1) I(>1且<4) R(≥2) R448K 支32(n=1) R(≥4) R(≥2) S83L, D530N, L553I, T570A, I702V, A735S, I743M, S789N, I790M, D794N, D799N, I831T 支42(n=1) I(>1且<4) R(≥2) S83L, D530N, L553I, T570A, I702V, A735S, I743M, S789N, I790M, D794N, D799N, I831T 支73(n=1) I(>1且<4) R(≥2) A136T, D530N, T570A, I702V, A734T, A735S, I743M, Q767E, S789N, I790M, D794N, D799N, M828I, I831T 支76(n=1) R(≥4) R(≥2) L176F S83L, D530N, L553I, T570A, I702V, A735S, I743M, S789N, I790M, D794N, D799N, I831T 支84/86(n=2) I(>1且<4) R(≥2) 尿17;支88/30(n=3) S(≤1) I(>1且<2) 支83/74/72(n=3) I(>1且<4) I(>1且<2) 支27(n=1) S(≤1) I(>1且<2) D530N, T570A, A735S, I743M, Q767E, S789N, I790M, D794N, D799N, I831T 支99(n=1) I(>1且<4) I(>1且<2) L540F 解脲脲原体 支6/39/50/91(n=4) R(≥4) R(≥2) S83L 支94(n=1) I(>1且<4) R(≥2) 支102/70;尿97(n=3) R(≥4) R(≥2) 尿36/40(n=2) S(≤1) R(≥2) 支37(n=1) S(≤1) I(>1且<2) A:丙氨酸Ala; R:精氨酸Arg; D:天冬氨酸Asp; C:半胱氨酸Cys; Q:谷氨酰胺Gln; E:谷氨酸Glu; H:组氨酸His; I:异亮氨酸Ile; G:甘氨酸Gly; N:天冬酰胺Asn; L:亮氨酸Leu; K:赖氨酸Lys; M:甲硫氨酸Met; F:苯丙氨酸Phe; P:脯氨酸Pro; S:丝氨酸Ser; T:苏氨酸Thr; W:色氨酸Trp; Y:酪氨酸Tyr; V:缬氨酸Val; R:耐药; I:中介; S:敏感; 下划线标注者为新发现突变 -
[1] Jorgensen JH, Pfaller MA. ISBN: 9781555817374. Manual of clinical microbiology[S]. 11th ed. Washington DC: ASM Press, 2015.
[2] Garcia LS. ISBN: 9781555815271. Clinical Microbiology Procedures Handbook[S]. 3rd ed. Washington DC: ASM Press, 2007.
[3] Zeng XY, Xin N, Tong XN, et al. Prevalence and antibiotic susceptibility of Ureaplasma urealyticum and Mycoplasma hominis in Xi'an, China[J]. Eur J Clin Microbiol Infect Dis, 2016, 35:1941-1947. DOI: 10.1007/s10096-016-2745-2
[4] Beeton ML, Chalker VJ, Kotecha S, et al. Comparison of full gyrA, gyrB, parC and parE gene sequences between all Ureaplasma parvum and Ureaplasma urealyticum serovars to separate true fluoroquinolones antibiotic resistance mutations from non-resistance polymorphism[J]. J Antimicrob Chemother, 2009, 64:529-538. DOI: 10.1093/jac/dkp218
[5] Beeton ML, Chalker VJ, Maxwell NC, et al. Concurrent titration and determination of antibiotic resistance in Ureaplasma species with identification of novel point mutations in genes associated with resistance[J]. Antimicrob Agents Chemother, 2009, 53:2020-2027. DOI: 10.1128/AAC.01349-08
[6] Lee MY, Kim MH, Lee WI, et al. Prevalence and Antibiotic Susceptibility of Mycoplasma hominis and Ureaplasma urealyticum in Pregnant Women[J]. Yonsei Med J, 2016, 57:1271-1275. DOI: 10.3349/ymj.2016.57.5.1271
[7] Redelinghuys MJ, Ehlers MM, Dreyer AW, et al. Antimicrobial susceptibility patterns of Ureaplasma species and Mycoplasma hominis in pregnant women[J]. BMC Infect Dis, 2014, 14:171. DOI: 10.1186/1471-2334-14-171
[8] 张睿, 叶阿里, 孔令君, 等.临床患者三种性传播疾病分子生物学检测分析[J].现代检验医学杂志, 2015, 30:107-110. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=sxyxjy201503031 [9] Song T, Ye A, Xie X, et al. Epidemiological investigation and antimicrobial susceptibility analysis of Ureaplasma species and Mycoplasma hominis in outpatients with genital manifestations[J]. J Clin Pathol, 2014, 67:817-820. DOI: 10.1136/jclinpath-2014-202248
[10] Piccinelli G, Gargiulo F, Biscaro V, et al. Analysis of mutations in DNA gyrase and topoisomerase Ⅳ of Ureaplasma urealyticum and Ureaplasma parvum serovars resistant to fluoroquinolones[J]. Infect Genet Evol, 2017, 47:64-67. DOI: 10.1016/j.meegid.2016.11.019
[11] Xiao L, Crabb DM, Duffy LB, et al. Chromosomal mutations responsible for fluoroquinolones resistance in Ureaplasma species in the United States[J]. Antimicrob Agents Chemother, 2012, 56:2780-2783. DOI: 10.1128/AAC.06342-11
[12] Bébéar CM, Renaudin H, Charron A, et al. DNA gyrase and topoisomerase Ⅳ mutations in clinical isolates of Ureaplasma spp. and Mycoplasma hominis resistant to fluoroquinolones[J]. Antimicrob Agents Chemother, 2003, 47:3323-3325. DOI: 10.1128/AAC.47.10.3323-3325.2003
[13] Duffy L, Glass J, Hall G, et al. Fluoroquinolones resistance in Ureaplasma parvum in the United States[J]. J Clin Microbiol, 2006, 44:1590-1591. DOI: 10.1128/JCM.44.4.1590-1591.2006
[14] Song J, Qiao Y, Kong Y, et al.Frequent topoisomerase Ⅳ mutations associated with fluoroquinolones resistance in Ureaplasma species[J]. J Med Microbiol, 2015, 64:1315-1320. DOI: 10.1099/jmm.0.000153
[15] Fernández J, Karau MJ, Cunningham SA, et al.Antimi-crobial Susceptibility and Clonality of Clinical Ureaplasma Isolates in the United States[J]. Antimicrob Agents Chemother, 2016, 60:4793-4798. DOI: 10.1128/AAC.00671-16
[16] Kamiya Y, Shimada Y, Ito S, et al. Analysis of the quinolones-resistance determining region of the gyrA gene and the analogous region of the parC gene in Ureaplasma parvum and Ureaplasma urealyticum detected in first-void urine of men with non-gonococcal urethritis[J]. J Antimicrob Chemother, 2013, 68:480-482. DOI: 10.1093/jac/dks417
[17] Kawai Y, Nakura Y, Wakimoto T, et al. In vitro activity of five quinolones and analysis of the quinolones resistance-determining regions of gyrA, gyrB, parC, and parE in Ureaplasma parvum and Ureaplasma urealyticum clinical isolates from perinatal patients in Japan[J]. Antimicrob Agents Chemother, 2015, 59:2358-2364. DOI: 10.1128/AAC.04262-14
[18] Schneider SC, Tinguely R, Droz S, et al. Antibiotic susceptibility and sequence type distribution of Ureaplasma species isolated from genital samples in Switzerland[J]. Antimicrob Agents Chemother, 2015, 59:6026-6031. DOI: 10.1128/AAC.00895-15
[19] 朱小飞, 彭红新, 李岷.喹诺酮抗性决定区域位点突变诱导解脲脲原体耐喹诺酮类药物的系统评价[J].临床检验杂志, 2014, 32:46-48. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=lcjyzz201501013 [20] Jin H, Qi C, Zou Y, et al. Biochanin A partially restores the activity of ofloxacin and ciprofloxacin against topoisomerase IV mutation-associated fluoroquinolones-resistant Ureaplasma species[J]. J Med Microbiol, 2017, 66:1545-1553. DOI: 10.1099/jmm.0.000598
[21] Robicsek AJ. Strahilevitz GA, Jacoby M, et al. Fluoroquinolones-modifying enzyme:a new adaptation of a common aminoglycoside acetyltransferase[J]. Nat Med, 2006, 12:83-88. DOI: 10.1038/nm1347
-
期刊类型引用(5)
1. 张兰,周竹娟,程畅,王玉涵,承文超,陈秀英,董开元,黄文. 重庆地区954例中枢神经系统感染性疾病的流行病学和临床特征分析. 解放军医学杂志. 2024(05): 534-541 .  百度学术
百度学术
2. 郭路明,于龙,李力韬,吴云峰,李大伟,鲍达,罗展鹏,刘宁,杨尚杰,崔旭. 宏基因组二代测序技术对结核性与非结核性脊柱感染疾病的诊断价值研究. 解放军医学院学报. 2024(05): 457-462+480 .  百度学术
百度学术
3. 户梦婷,张栋,贾沛瑶,陆旻雅,周梦兰,郭佳钰,苏慧婷,高羿,席婧媛,朱华栋,杨启文. 生食感染广州管圆线虫蜗牛引起嗜酸性粒细胞增多性脑膜炎1例. 协和医学杂志. 2024(06): 1463-1467 .  本站查看
本站查看
4. 张永杰,王兴华,黄晓岚. 不同抗菌药物方案在中枢神经系统感染性疾病治疗中的应用效果比较. 医药前沿. 2024(29): 28-30+34 .  百度学术
百度学术
5. 尚远江,邾国庆,张蕾,沈丹丹,潘秋辉. 宏基因组二代测序和传统方法在中枢神经系统感染中的临床应用. 检验医学. 2024(11): 1101-1107 .  百度学术
百度学术
其他类型引用(2)

 作者投稿
作者投稿 专家审稿
专家审稿 编辑办公
编辑办公 邮件订阅
邮件订阅 RSS
RSS

 下载:
下载: