Diversity Analysis of Intestinal Microbiota in Psoriasis Patients: A Single-center Prospective Study
-
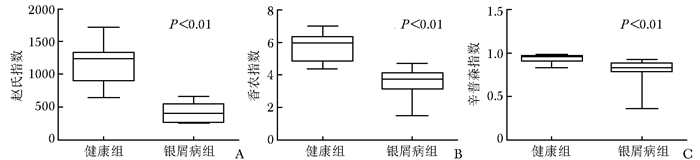
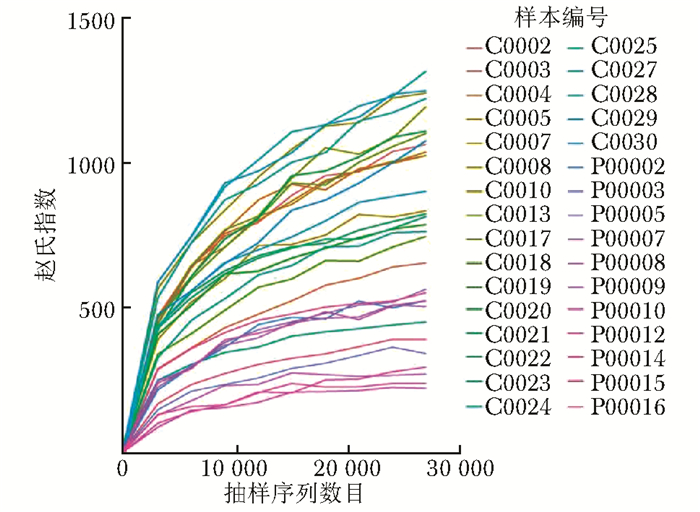
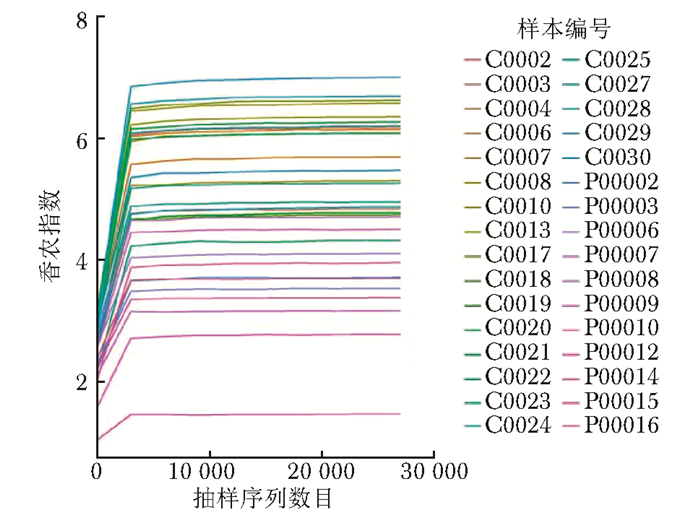
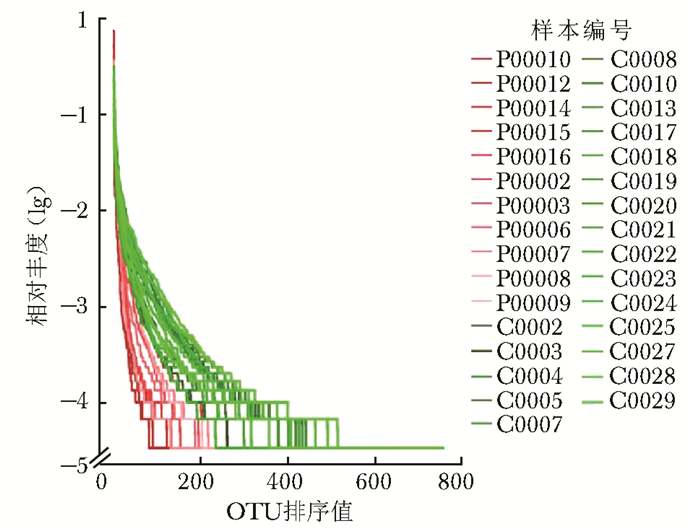
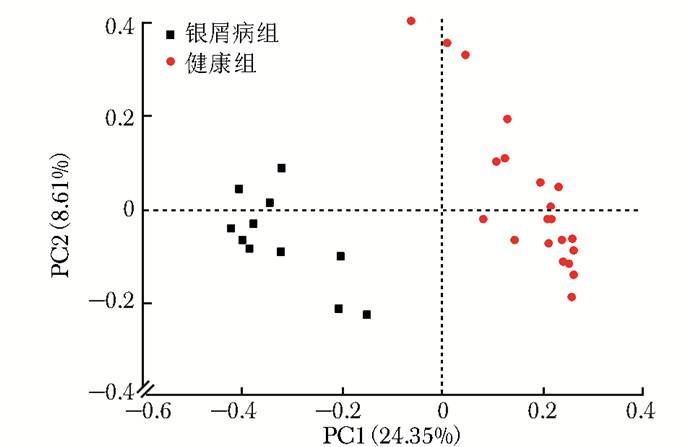
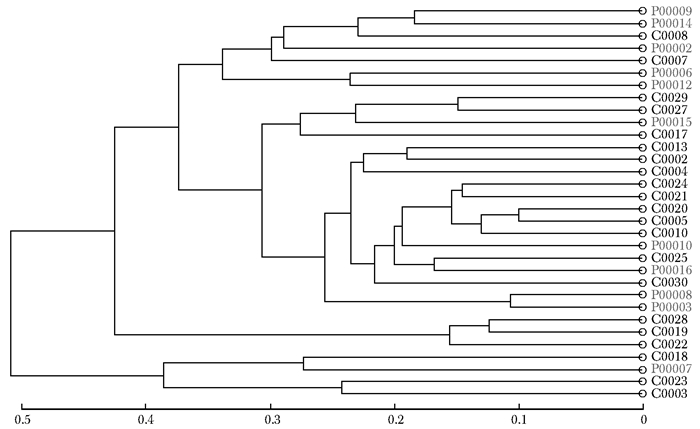
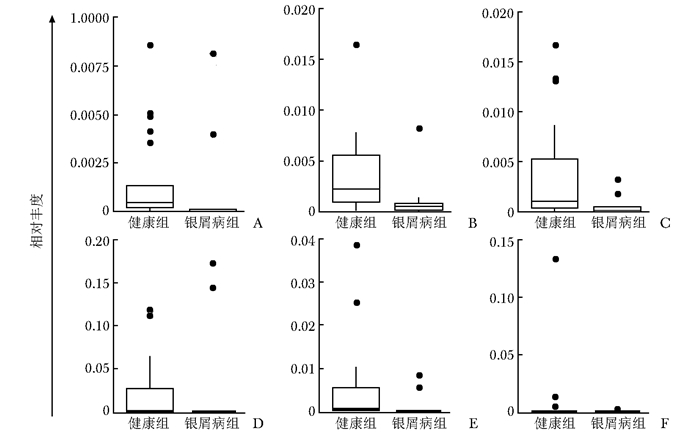
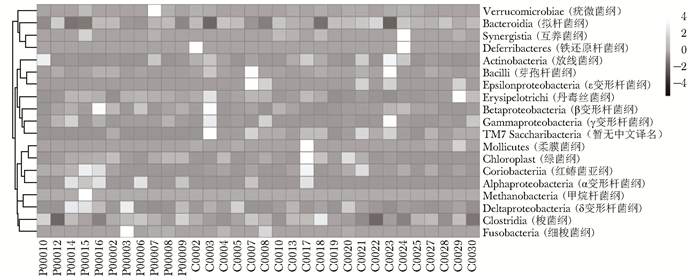
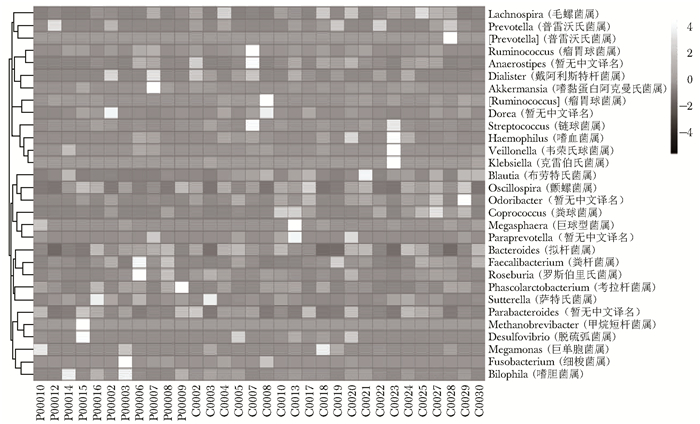
摘要:目的 探讨银屑病患者与健康人群肠道菌群多样性的差异。方法 收集2017年5月至2018年6月间在中国医学科学院皮肤病研究所住院治疗的银屑病患者(银屑病组)及同期本院健康体检人群(健康组)的新鲜粪便标本, 分析受试者相关临床资料。提取肠道菌群DNA, 采用16S rDNA基因扩增和Illumina平台双端2×300策略测序, 基于Gold数据库按>97%的相似性聚类操作分类单元(operational taxonomic unit, OTU), 对照Silva数据库进行物种注释及分类, 各层级样本采用轶和检验分析物种差异; QIIME软件计算α多样性主要指数、β多样性分析, t检验分析指数差异, P<0.05为差异有统计学意义。相关研究结果通过R及GraphPad Prism作图展示。结果 符合入选和排除标准的11例银屑病患者及21例健康对照者入选本研究, 两组研究对象的性别构成比、年龄和体质量指数无统计学差异(P均>0.05)。DNA测序分析显示样本测序覆盖深度>0.99。银屑病组肠道菌群的OTU数量(278.18±89.75比722.95±152.81, t=10.36, P<0.01)、赵氏指数(433.38±147.47比1156.08±292.50, t=9.291, P<0.01)、香农指数(3.56±0.87比5.73±0.78, t=6.972, P<0.01)和辛普森指数(0.79±0.15比0.94±0.04, t=3.287, P<0.01)均显著低于健康组。Rank-Abundance曲线显示银屑病组肠道菌群均匀程度较低。PCoA分析(unweighted)显示银屑病组与健康组在第一主成分(24.35%)可显著分离, Weighted UniFrac分析可见银屑病组混杂在健康样本中无法区分, 且与银屑病亚型无关。样本聚类分析显示, 银屑病组肠道菌群与健康组有一定重叠性, 银屑病样本特异性菌群少; 在门层级, 健康组中可检测到TM7, 相对丰度为0.000 066 9(0.000 033 4~0.000 200 5), 而银屑病组仅在个别样本中显示有微量存在, 相对丰度为0(P<0.05);在属层级, 银屑病组双歧杆菌属[0.000 033 4(0.000 016 7~0.000 100 3)比0.000 401 1(0.000 200 5~0.001 337 0)]、布劳特氏菌属[0.000 467 9(0.000 183 8~0.000 434 5)比0.002 206 0(0.000 935 9~0.005 582 0)]、粪球菌属[0.000 033 4(0~0.000 401 1)比0.000 902 5(0.000 334 2~0.005 315 0)]较健康组显著降低, 健康组中的戴阿利斯特杆菌属和嗜血菌属仅在银屑病组个别样本中微量存在, 克雷伯氏杆菌属在健康组和银屑病组个别样本中存在, 但在银屑病组的相对丰度更低, 接近0(P均<0.05)。个体样本高丰度菌群分析显示, 银屑病组个别样本中多糖及短链脂肪酸代谢相关菌群的相对丰度降低。结论 银屑病患者肠道菌群多样性低于健康人群。Abstract:Objective The aim of this study was to investigate the difference in the diversity of intestinalmicrobiota of psoriasis patients compared with that of healthy people.Methods Fresh fecal samples and clinical data of hospitalized psoriasis patients from May 2017 to June 2018 in the Institute of Dermatology of Chinese Academy of Medical Sciences & Peking Union Medical College were prospectively collected. Healthy people undergoing physical examination during the same period were selected as the healthy control. After DNA extraction of intestinal microbiota and the gene amplification of 16S rDNA, the paired-end 2×300 strategy was used to sequence by Illumina platform. According to Gold database and the similarity-clustering operational taxonomic unit (OUT) > 97%, intestinal microbiota was annotated and classified by Silva database. The species difference of samples at all levels was analyzed using Rank sum test. The main index of α diversity and the analysis of β diversity were calculated using QIIME software. All indexes were analyzed by Welch's t test and P < 0.05 was considered statistically significant. The results were shown by R and GraphPad Prism plotting.Results Totally 11 psoriasis patients and 21 healthy controls were eligible for this study. Between the psoriasis group and the healthy group, the ratios of gender, age, and body mass index showed no statistical significance (all P > 0.05). The DNA sequencing showed that coverage index of samples' sequencing was > 0.99. The OTU numbers (278.18±89.75 vs. 722.95±152.81, t=10.36, P < 0.01), Zhao index (433.38±147.47 vs. 1156.08±292.50, t=9.291, P < 0.01), Shannon index (3.56±0.87 vs. 5.73±0.78, t=6.972, P < 0.01), and Simpson index (0.79±0.15 vs. 0.94±0.04, t=3.287, P < 0.01) of the particular species of intestinal microbiota in the psoriasis group were significantly lower than those of the healthy group. The Rank-abundance curve showed that the psoriasis group had a lower evenness of intestinal microbiota. PCoA analysis (unweighted) showed that the first principal component (24.35%) was significantly separated between the two groups; however, Weighted UniFrac analysis showed that it was not clearly distinguished when the samples of the psoriasis group were mixed with the healthy group, which was independent of the subtypes of psoriasis. Through the sample clustering analysis, the intestinal microbiota partly overlapped between the two groups and the special microbiota were less in the psoriasis group. In the phylum level, TM7 could be detected in the healthy group and its relative abundance was 0.000 066 9 (0.000 033 4~0.000 200 5), while it was at micro-level in a few samples of the psoriatic group with a relative abundance of 0(Rank sum test, P < 0.05). On the genus level, the psoriasis group had significantly lower bifidobacterium[0.000 033 4(0.000 016 7~0.000 100 3) vs. 0.000 401 1 (0.000 200 5~0.001 337 0)], brauteria[0.000 467 9 (0.000 183 8~0.000 434 5) vs. 0.002 206 0(0.000 935 9~0.005 582 0)], and fecal coccus[0.000 033 4 (0~0.000 401 1) vs. 0.000 902 5(0.000 334 2~0.005 315 0)] compared to the healthy group. Dialisteria and haemophilus appearing in the healthy group were only found in a few samples in the psoriasis group. Klebsiella existed in a few samples of the two groups but was lower in the psoriasis group (relative abundance was close to 0) (Rank sum test, all P < 0.05). The high abundance analysis of individual samples showed that in some psoriasis samples, the abundance of the taxa related to the metabolism of polysaccharides and short-chain fatty acids was decreased.Conclusion Diversity of intestinal microbiota in psoriasis patients is lower than that of healthy people.
-
Keywords:
- psoriasis /
- intestinal microbiota /
- 16S rDNA /
- sequencing
-
肺血栓栓塞症(pulmonary thromboembolism, PTE)发病率为100~200/10万[1],在心血管疾病中仅次于冠状动脉粥样硬化性心脏病和高血压,由于中国人口基数庞大,故PTE患者数量可观。PTE评估指标包括短期死亡率、复发率以及治疗相关并发症(包括慢性血栓栓塞性肺动脉高压等),除此之外,对PTE患者的生活质量评估也是随诊评估的重要组成部分。
通过构建多维度体系评估疾病和治疗对患者生理、心理、社会功能、幸福感、总体健康状况的影响,对评定疾病预后具有重要价值。较低生活质量评分与疾病预后不佳及生存率降低有关[2-3],而治疗措施本身也可直接影响生活质量评分中的若干项指标。本研究通过健康状况调查问卷SF-36量表来评定PTE患者的生活质量,观察PTE对患者生活质量的影响,并通过单因素和多因素回归分析探讨影响患者生活质量的相关因素。
1. 对象与方法
1.1 研究对象
2016年5月至2017年11月所有在北京协和医院住院治疗或呼吸内科门诊就诊的PTE患者,符合入选和排除标准者应邀填写SF-36量表,在门诊或病房现场填写,或通过网页、电子邮件及邮寄等方式获取调查问卷。
1.1.1 入选标准
(1) 患者年龄>18周岁;(2)通过CT肺动脉造影或核素通气灌注显像确诊PTE;(3)病情稳定,可独立完成问卷调查。
1.1.2 排除标准
(1) 无法沟通、不愿参加问卷调查或不能提供详细病史者;(2)为减少急性发作对生活质量评估的影响,诊断PTE在1个月内者;(3)为降低并发症影响,明确诊断慢性血栓性肺动脉高压者;(4)治疗期间出现肺栓塞复发者。
1.2 生活质量评估
采用SF-36量表(中文版)评估患者生活质量。SF-36量表是美国波士顿健康研究所研制的简明健康调查问卷,广泛用于普通人群生存质量测定、临床试验效果评价以及卫生政策评估等领域。SF-36量表作为简明健康调查问卷,包括生理机能(physical functioning, PF)、生理职能(role-physical, RP)、躯体疼痛(bodily pain, BP)、一般健康状况(general health, GH)、精力(vitality, VT)、社会功能(social functioning, SF)、情感职能(role-emotional, RE)、精神健康(mental health, MH)和健康变化(health transition, HT)等9个变量,全面反映被调查者的生存质量。
1.3 研究内容
计算SF-36量表 9个维度的得分情况并以百分制折算,与中国普通人群(数据来自北京、广州、苏州、重庆、郑州和大连的随机抽取样本住户中年满14周岁、具有阅读能力的全体住户成员)中各维度基线评分[4]对比,以明确PTE对患者生活质量的影响。
纳入可能与生活质量有关的11项临床资料,包括性别、年龄、肥胖、间隔时间(诊断PTE时间至填表时间)、栓塞部位、深静脉血栓、癌症、慢性心肺疾病、诱发型PTE、一过性危险因素及永久性危险因素,与SF-36量表各维度评分进行相关分析,探讨影响生活质量评分的相关因素。
1.4 统计学处理
采用IBM SPSS Statistics 22软件进行统计学分析。计量资料结果以均数±标准差表示,计数资料结果以百分比表示。由于SF-36量表在中国人群的基线资料只有均数±标准差,缺乏具体样本例数,因此我们通过SPSS语法编辑器编程完成与本研究中数据的t检验[5]。随后将临床上可能的相关因素与SF-36量表中9个变量逐一进行单因素线性回归分析,以确定影响生活质量的临床因素;同时将所有可能的临床因素纳入多元线性回归分析,采取逐步回归法确定独立相关因素。判定系数(r2)用于评估回归对实际数据的拟合程度优劣。P<0.05为差异有统计学意义。
2. 结果
2.1 一般情况
共收集调查问卷103份,88份符合纳入和排除标准的PTE患者问卷进入最终分析。入选患者平均年龄(53±16)岁,男女比例为1:1.2,体质量指数(body mass index,BMI)为(25.7±4.4) kg/m2,间隔时间为(10.3±16.3)个月。中心性血栓(主干或主肺动脉)占45.5%(40/88), 深静脉血栓占50.0%(44/88),恶性肿瘤患者占15.9%(14/88),慢性心肺疾病患者占44.3%(39/88),复发性PTE占4.5%(4/88),非诱发型PTE占25.0%(22/88),而一过性危险因素患者占29.5%(26/88)。
2.2 SF-36量表评分情况
PTE患者SF-36量表各维度评分介于30.4~60.1之间,其中得分最低的变量是生理职能,其次为健康变化、一般健康状况与情感职能(表 1)。除精神健康变量外,其他8个维度PTE发生1年以上组的生活质量评分均高于1年以内组,其中生理职能变量具有统计学差异(P<0.01)。和普通人群的生活质量评分相比,PTE患者各维度评分均低于普通人群(表 2),均有统计学差异(P均<0.01),其中生理职能评分差异最大(52.6),精力评分差异最小(12.8)。
表 1 PTE患者SF-36量表各维度评分维度 全部PTE患者(n=88) PTE≤1年(n=73) PTE>1年(n=15) P值 生理机能 59.8±27.4 57.5±26.7 71.0±28.4 0.0811 生理职能 30.4±41.9 25.7±39.5 53.3±47.1 0.0193 躯体疼痛 67.4±23.6 65.3±24.1 77.5±18.8 0.0684 一般健康状况 43.0±23.1 41.3±22.6 51.3±24.5 0.1275 精力 57.3±23.5 55.8±22.4 64.3±28.3 0.2047 社会功能 60.1±29.9 58.4±29.9 68.1±30.0 0.2559 情感职能 44.7±43.1 41.6±42.6 60.0±44.0 0.1333 精神健康 58.7±21.5 58.9±21.6 57.9±22.1 0.8711 健康变化 37.8±36.2 35.3±37.0 50.0±29.9 0.1527 PTE:肺血栓栓塞症 表 2 PTE患者SF-36量表各维度评分及与中国普通人群的比较维度 中国普通人群[4]
(n=4251)PTE患者
(n=88)差值 P值 生理机能 87.6±16.8 59.8±27.4 27.8 <0.01 生理职能 83.0±20.7 30.4±41.9 52.6 <0.01 躯体疼痛 83.3±19.7 67.4±23.6 15.9 <0.01 一般健康状况 68.2±19.4 43.0±23.1 25.2 <0.01 精力 70.1±16.8 57.3±23.5 12.8 <0.01 社会功能 84.8±16.6 60.1±29.9 24.7 <0.01 情感职能 85.3±17.7 44.7±43.1 40.6 <0.01 精神健康 78.8±15.4 58.7±21.5 20.1 <0.01 健康变化 - 37.8±36.2 - - PTE:同表 1 2.3 单因素回归分析
单因素回归分析显示,年龄增加与一般健康状况评分降低相关(P=0.026),间隔时间则与躯体疼痛正相关(P=0.017);诱发型PTE患者的一般健康状况评分低于非诱发型PTE患者(P=0.048);一过性危险因素导致的PTE与多项生活质量维度评分降低相关,包括生理机能(P=0.004)、精力(P=0.033)、社会功能(P=0.007)、精神健康(P=0.008)和健康变化(P=0.020),而永久性危险因素导致的PTE与精神健康正相关(P=0.006)。
2.4 多因素回归分析
对性别、年龄、肥胖(体质量指数>28 kg/m2)、间隔时间、栓塞部位、DVT、癌症、慢性心肺疾病、诱发型PTE、一过性高危因素、永久性高危因素等11项人口学及临床因素行多因素线性回归分析,结果显示生理机能、躯体疼痛、一般健康状况、精力、社会功能、精神健康和健康变化这7项变量可以通过回归模型描述。年龄增加与一般健康状况评分降低相关(非标准化回归系数为-0.37±0.15,P=0.013);间隔时间与躯体疼痛评分正相关(非标准化回归系数为0.39±0.16,P=0.017);诱发型PTE患者的一般健康状况评分低于非诱发型PTE患者(非标准化回归系数为-13.0±5.5,P=0.024);一过性危险因素导致的PTE与多项维度评分降低相关,包括生理机能(非标准化系数为-18.0±6.1,P=0.004)、精力(非标准化系数为-12.0±5.3,P=0.033)、社会功能(非标准化系数为-19.0±6.7,P=0.007)和健康变化(非标准化系数为-20.0±8.2,P=0.020);永久性危险因素导致的PTE与精神健康正相关(非标准化系数为12.6±4.4,P=0.006)。与单因素回归分析结果相比,仅精神健康项略有不同,其他维度大致相同。
在包含纳入因素的线性回归模型中,相关因素对解释维度评分变化的力度较弱,仅有4.1%~9.1%(r2)的维度变化可使用独立相关因素加以解释,其中精力最低(r2=4.1%), 一般健康状况最高(r2=9.1%),生理机能、躯体疼痛、社会功能、精神健康、健康变化的r2值分别为8.3%、5.3%、7.0%、7.5%和5.0%。
3. 讨论
本研究分析了88例PTE患者的生活质量评分情况,并与中国普通人群比对,结果显示,PTE患者的生活质量各维度评分均显著低于普通人群,提示PTE患者的生理功能、心理功能与社会活动各方面均受到明显限制,同时发现多项与PTE相关的临床及人口学因素可独立影响生活质量评分。
迄今为止,仅有几项研究系统分析了PTE对患者生活质量的影响,国内有关PTE生活质量的评估研究亦少见。Klok等[6]的研究显示,PTE患者SF-36量表中的8个维度(未纳入健康变化变量)评分均低于正常人群;而van Es等[7]的研究中只有社会功能、生理职能、情感职能、精力和一般健康状况5个维度低于正常人群;Tavoly等[8]采用EQ-5D健康量表和视觉模拟标尺评分评估PTE患者的生活质量,同样发现PTE患者的生活质量低于正常人群;一些荟萃分析显示PTE患者的生理健康总分低于正常人群[9-10];对中国急性PTE患者进行的另一项调查同样显示PTE患者的生理健康和心理健康评分显著低于正常人群[13]。
并非所有研究均获得以上类似结果,在ELOPE前瞻性队列研究中,PTE患者在1个月时的SF-36量表评分显著低于正常人群,但在12个月时却无差别[11-12]。本研究显示,中国PTE患者SF-36量表中所有维度评分均低于正常人群,且评分绝对值和评分与正常人群差值均较Klok和van Es的结果低,这可能与纳入患者的基础疾病及间隔时间等因素有关。间隔时间超过1年PTE患者的生活质量评分普遍高于1年内组,说明间隔时间对评分具有重要影响。但在校正间隔时间等因素后,本研究中多个维度,如生理职能、社会功能、情感职能和精神健康等的评分依然显著低于Klok的研究,提示除本文涉及的临床因素外,种族、文化等多方面因素也可能对PTE患者生活质量产生影响。
本研究中年龄与一般健康状况呈负相关,即年龄越大,一般健康状况越差,其他临床特征,如性别、体质量指数、癌症、慢性心肺疾病等均未发现与生活质量相关;PTE专属特征中的间隔时间与躯体疼痛评分正相关,说明间隔时间越长,躯体疼痛越轻(评分越高);诱发型PTE患者的一般健康状况评分低于非诱发型,可能与诱发型患者合并其他基础疾病影响了生活质量评分有关。本研究中一过性危险因素对生活质量的影响最大,它同时与生理机能、精力、社会功能、健康变化呈负相关,而永久性危险因素则与精神健康正相关,提示一过性危险因素相关的PTE患者生活质量评分低于永久性危险因素患者,这一结果不同于一般认知,可能与一过性危险因素患者对生活质量预期较高有关,但不能确定是否与一过性危险因素患者抗凝治疗时间整体偏短有关,需进一步研究。PTE部位及严重程度并不影响生活质量评分,这与Klok研究结果相似,之前一项针对深静脉血栓的研究亦提示血栓部位及严重程度不影响患者生活质量[14]。
本研究中各维度的判定系数r2<10%,说明这些相关因素模型尚不能准确解释生活质量评分变化,可能与SF-36量表属广义评分表,并非专门针对PTE设计相关。目前已有专门针对PTE患者生活质量评分的问卷(PEmb-Qol)[15-16],其是否适用于中国PTE患者尚需进一步验证。此外,其他尚未纳入本研究的因素,如受教育程度、收入、社会地位、地理分布、疾病认知、行为应对、自我管理等个体化因素均可能影响生活质量评分,在将来的扩大研究中有必要将这些潜在相关因素纳入生活质量回归分析。
本研究存在一些不足之外。首先,虽然本研究纳入的患者例数较国内类似研究多,但与Klok研究相比,本研究纳入患者数量仍相对较少,由于潜在相关因素较多,可能会对结果产生一定影响。其次,由于老年患者受教育程度普遍偏低,大多数无法独立完成该项问卷调查,因此被排除在本调查之外,从而导致研究对象存在一定偏倚。再次,本研究中患者平均年龄较中国正常人群偏高,在比对这两组结果时年龄是一项混杂因素。最后,由于样本量不够大,本研究中某些维度呈非正态分布,但为了便于和普通人群的均值对比,仍沿用了均数±标准差的表述方式,其对比结果可能会存在一定误差。
本研究深层次探索了PTE对中国人群生活质量的影响,对相关影响因素进行全面评估,并发现数个影响生活质量的独立相关因素,为将来进一步探索PTE患者生活质量与治疗及预后的相关性提供了初步线索。
利益冲突 无 -
图 3 银屑病患者与健康人群肠道菌群的香农指数曲线
C、P:同图 2
图 4 银屑病患者与健康人群肠道菌群的Rank-Abundance曲线
横坐标为OTU按丰度(序列条数)由大到小等级排序,纵坐标为每个OTU等级中所含序列数的相对丰度; C、P:同图 2; OTU:分类操作单元
图 6 银屑病患者与健康人群肠道菌群的Weighted UniFrac分析
C、P:同图 2
-
[1] 吴超, 晋红中.银屑病的危险因素和流行分布[J].协和医学杂志, 2012, 3:471-475. DOI: 10.3969/j.issn.1674-9081.2012.04.024 [2] Zakostelska Z, Malkova J, Klimesova K, et al. Intestinal microbiota promotes psoriasis-like skin inflammation by enhancing Th17 response[J]. PLoS One, 2016, 11:e159539. http://europepmc.org/articles/PMC4951142/
[3] Skov L, Baadsgaard O. Bacterial superantigens and inflammatory skin diseases[J]. Clin Exp Dermatol, 2000, 25:57-61. DOI: 10.1046/j.1365-2230.2000.00575.x
[4] Fry L, Baker BS. Triggering psoriasis:the role of infections and medications[J]. Clin Dermatol, 2007, 25:606-615. DOI: 10.1016/j.clindermatol.2007.08.015
[5] Scarpa R, Manguso F, D'Arienzo A, et al. Microscopic inflammatory changes in colon of patients with both active psoriasis and psoriatic arthritis without bowel symptoms[J]. J Rheumatol, 2000, 27:1241-1246. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=a79555c858f4a1cd0f1759c3afa2de15
[6] Shen S, Wong CH. Bugging inflammation:role of the gut microbiota[J]. Clin Transl Immunology, 2016, 5:e72. DOI: 10.1038/cti.2016.12
[7] Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography[J]. Nature, 2012, 486:222-227. DOI: 10.1038/nature11053
[8] Scher JU, Ubeda C, Artacho A, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease[J]. Arthritis Rheumatol, 2015, 67:128-139. DOI: 10.1002/art.38892
[9] Eppinga H, Sperna WC, Thio HB, et al. Similar depletion of protective faecalibacterium prausnitzii in psoriasis and inflammatory bowel disease, but not in hidradenitis suppurativa[J]. J Crohns Colitis, 2016, 10:1067-1075. DOI: 10.1093/ecco-jcc/jjw070
[10] Brusca SB, Abramson SB, Scher JU. Microbiome and mucosal inflammation as extra-articular triggers for rheumatoid arthritis and autoimmunity[J]. Curr Opin Rheumatol, 2014, 26:101-107. DOI: 10.1097/BOR.0000000000000008
[11] Khanna S, Tosh PK. A clinician's primer on the role of the microbiome in human health and disease[J]. Mayo Clin Proc, 2014, 89:107-114. DOI: 10.1016/j.mayocp.2013.10.011
[12] Sommer F, Backhed F. The gut microbiota-masters of host development and physiology[J]. Nat Rev Microbiol, 2013, 11:227-238. DOI: 10.1038/nrmicro2974
[13] Brinig MM, Lepp PW, Ouverney CC, et al. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease[J]. Appl Environ Microbiol, 2003, 69:1687-1694. DOI: 10.1128/AEM.69.3.1687-1694.2003
[14] Kuehbacher T, Rehman A, Lepage P, et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease[J]. J Med Microbiol, 2008, 57:1569-1576. DOI: 10.1099/jmm.0.47719-0
[15] Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota[J]. Science, 2013, 341:1237439. DOI: 10.1126/science.1237439
[16] Shin NR, Whon TW, Bae JW. Proteobacteria:microbial signature of dysbiosis in gut microbiota[J]. Trends Biotechnol, 2015, 33:496-503. DOI: 10.1016/j.tibtech.2015.06.011
[17] Caesar R, Tremaroli V, Kovatcheva-Datchary P, et al. Crosstalk between gut microbiota and dietary lipids aggra-vates WAT inflammation through TLR signaling[J]. Cell Metab, 2015, 22:658-668. DOI: 10.1016/j.cmet.2015.07.026
[18] Feng Z, Long W, Hao B, et al. A human stool-derived Bilophila wadsworthia strain caused systemic inflammation in specific-pathogen-free mice[J]. Gut Pathog, 2017, 9:59. DOI: 10.1186/s13099-017-0208-7
-
期刊类型引用(2)
1. 吴晔,王寅冰. 高流量鼻导管吸氧联合精细化护理在低氧性呼吸衰竭患者中的应用. 国际医药卫生导报. 2021(04): 596-599 .  百度学术
百度学术
2. 陈静媚,黄小芸,蒋丽梅. 专科护理在门诊痛风患者治疗中的应用. 当代护士(中旬刊). 2019(12): 97-100 .  百度学术
百度学术
其他类型引用(1)

 作者投稿
作者投稿 专家审稿
专家审稿 编辑办公
编辑办公 邮件订阅
邮件订阅 RSS
RSS

 下载:
下载: